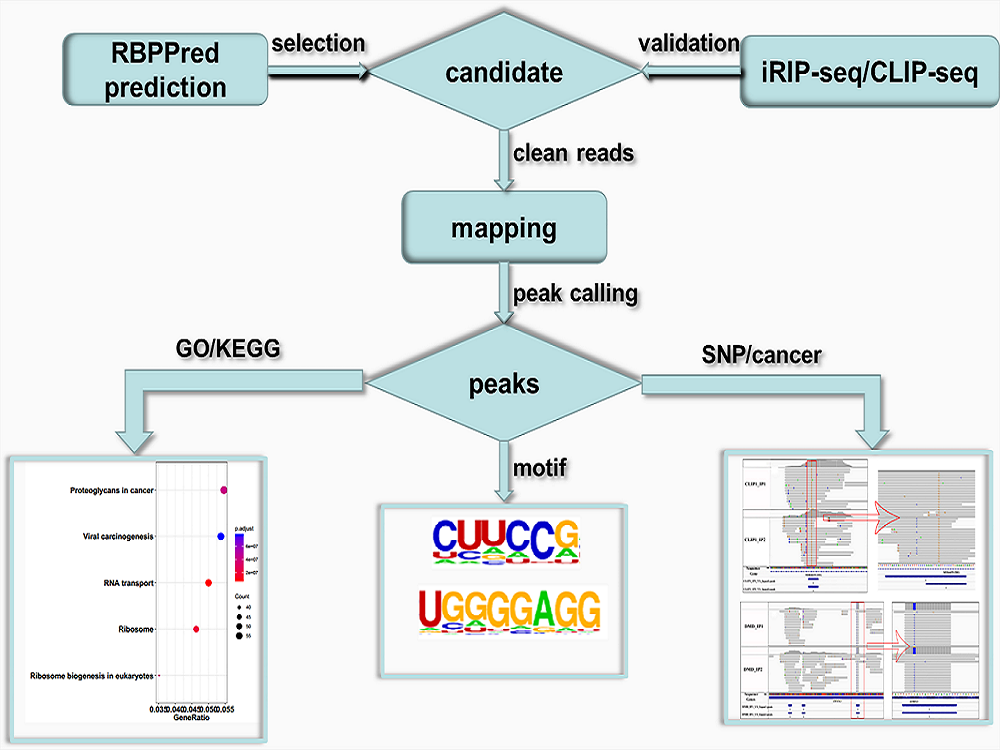
The requested programs:
Cutadapt: A software for removing adapters(Version 1.12,2016-11-28). For more information, please see https://cutadapt.readthedocs.io/en/stable/installation.html
or you can type"pip install cutadapt==1.12" to install cutadapt
FASTX-Toolkit: It is a collection of command line tools for Short-Reads FASTA/FASTQ files preprocessing (Version 0.0.13,2010-02-02). It can be downloaded from http://hannonlab.cshl.edu/fastx_toolkit/fastx_toolkit-0.0.13.tar.bz2 For more information, please see http://hannonlab.cshl.edu/fastx_toolkit/download.html
or you can type:
wget http://hannonlab.cshl.edu/fastx_toolkit/fastx_toolkit_0.0.13_binaries_Linux_2.6_amd64.tar.bz2
tar -xjf fastx_toolkit_0.0.13_binaries_Linux_2.6_amd64.tar.bz2
cp ./bin/* $HOME/bin
BEDTools: A powerful toolset for genome arithmetic. (Version 2.20.1). It can be downloaded from https://github.com/arq5x/bedtools2/releases/download/v2.20.1/bedtools-2.20.1.tar.gz For more information, please see https://github.com/arq5x/bedtools2
or you can type:
wget https://github.com/arq5x/bedtools2/releases/download/v2.20.1/bedtools-2.20.1.tar.gz
tar -zxvf v2.20.1.tar.gz
cd bedtools2-2.20.1
make
cp bin/* $HOME/bin
Bowtie2: It is an ultrafast and memory-efficient tool for aligning sequencing reads to long reference sequences. (Version 2.2.5). It can be downloaded from https://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.2.5/bowtie2-2.2.5-source.zip/download
after download, you can type:
unzip bowtie2-2.2.5-source.zip
cd bowtie2-2.2.5
make
cp bowtie2* $HOME/bin
TopHat2: It is a fast splice junction mapper for RNA-Seq reads (Version 2.1.1,2016-02-23). It can be downloaded from https://ccb.jhu.edu/software/tophat/downloads/tophat-2.1.1.Linux_x86_64.tar.gz For more information, please see https://ccb.jhu.edu/software/tophat/index.shtml
or you can type:
wget https://ccb.jhu.edu/software/tophat/downloads/tophat-2.1.1.Linux_x86_64.tar.gz
tar zxvf tophat-2.1.1.Linux_x86_64.tar.gz
cd tophat-2.1.1.Linux_x86_64/
cp -r * $HOME/bin
HTSlib: A C library for reading/writing high-throughput sequencing data (Version 1.9). It can be downloaded from https://github.com/samtools/htslib/releases/download/1.9/htslib-1.9.tar.bz2 For more information, please see http://www.htslib.org/
or you can type:
wget https://github.com/samtools/htslib/releases/download/1.9/htslib-1.9.tar.bz2
tar -jxvf htslib-1.9.tar.bz2
cd htslib-1.9
./configure --prefix=$HOME
make & make install
SAMtools: It is a suite of programs for interacting with high-throughput sequencing data. (Version 1.8,2018-04-03). It can be downloaded from https://sourceforge.net/projects/samtools/files/samtools/1.8/samtools-1.8.tar.bz2/download For more information, please see http://samtools.sourceforge.net/
after download, you can type:
tar -jxvf samtools-1.8.tar.bz2
cd samtools-1.8
./configure --prefix=$HOME --with-htslib="the path of htslib-1.9"
make & make install
HOMER: It is a suite of tools for Motif Discovery and next-gen sequencing analysis (Version 4.8.2). It can be downloaded from http://homer.ucsd.edu/homer/data/software/homer.v4.8.2.zip. For more information, please see http://homer.ucsd.edu/homer/. Please refer to the "README.txt" in the homer folder for the installation method.
gencode.v23.annotation.gff3 and GRCh38.p3.genome.fa: Reference genomic data (Version 23). It can be downloaded from ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_23/
novoalign: It is used for mapping(V3.07.00). It can be downloaded from http://www.novocraft.com/support/download/
after download, you can type:
Add the novocraft directory to your executable path
i.e. edit your ~/.bash_profile file to include:
PATH=$PATH:/home/novocraft
build the index of novoalign by typing:
./novoindex -k 14 -s 1 GRCH38_gencode_v23.ndx GRCh38.p3.genome.fa
or you can download the index of novoalign from
http://www.rnabinding.com/phdRBP/data/GRCH38_gencode_v23.ndx
CTK: It provides a set of tools for analysis of CLIP data starting from the raw reads generated by the sequencer (Version 1.0.3,2016-08-08). It can be downloaded from https://zhanglab.c2b2.columbia.edu/index.php/CTK_Documentation
or you can type:
1. wget https://cpan.metacpan.org/authors/id/C/CA/CALLAHAN/Math-CDF-0.1.tar.gz
tar -zxvf Math-CDF-0.1.tar.gz
cd Math-CDF-0.1
perl Makefile.PL
make & make install
2. wget https://github.com/chaolinzhanglab/ctk/archive/v1.0.7.tar.gz
tar -zxvf v1.0.7.tar.gz
Add the ctk directory to your executable path:
i.e. edit your ~/.bash_profile file to include:
export PATH=$PATH:$HOME/ctk-1.0.7
Piranha: It is a tool developed for peak calling (Version 1.2.1). It can be downloaded from http://smithlabresearch.org/downloads/piranha-1.2.1.tar.gz
or you can type:
1. wget http://mirrors.kernel.org/gnu/gsl/gsl-2.2.tar.gz
tar -zxvf gsl-2.2.tar.gz
cd gsl-2.2
./configure --prefix=$HOME/gsl
make & make install
Add the gsl directory to your executable path:
i.e. edit your ~/.bash_profile file to include:
export C_INCLUDE_PATH=$C_INCLUDE_PATH:$HOME/gsl/include
export CPLUS_INCLUDE_PATH=$CPLUS_INCLUDE_PATH:$HOME/gsl/include
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH::$HOME/gsl/lib
export LIBRARY_PATH=$LIBRARY_PATH::$HOME/gsl/lib
2. wget http://smithlabresearch.org/downloads/piranha-1.2.1.tar.gz
cd piranha-1.2.1
./configure --prefix=$HOME
make & make install
Add the Piranha directory to your executable path:
i.e. edit your ~/.bash_profile file to include:
export PATH=$PATH:$HOME/piranha/piranha-1.2.1/bin
Contact us:
Any questions about phdRBP, please email to liushiyong@gmail.com.
Reference:
Juan Xie, Xiaoli Zhang, Jinfang Zheng, Xu Hong, Xiaoxue Tong, Xudong Liu, Yaqiang Xue, Xuelian Wang, Yi Zhang and Shiyong Liu
Two novel RNA-binding proteins identification through computational prediction and experimental validation.
Genomics, S0888-7543(21)00429-8, 15 December 2021
Last modified: Fri. Oct. 30 10:32:00 CST 2020